tl;dr
You can convert a line drawing to datapoints with a sprinkle of {magick}.
Ape escape
Have you seen that video where you’re so focused on counting basketball passes that you fail to see the gorilla moving across the screen?
This kind of selective attention was studied by two researchers, Yanai and Lercher, who provided subjects with a dataset that looked like a gorilla when plotted. The gorilla was found less often if the subjects were also given a hypothesis to investigate.
The study got some attention on Twitter last week. As a result, Isabella Velásquez wrote a great blogpost where she recreated the dataset using R and Python in tandem via the {reticulate} package.
I had a go at creating the dataset with base R and the excellent {magick} package for image manipulation.
Point it out
The jpeg image file used in the original paper can be downloaded from classroomclipart.com to a temporary location on your machine.
download.file(
paste0(
"https://classroomclipart.com/images/gallery/",
"Clipart/Black_and_White_Clipart/Animals/",
"gorilla-waving-cartoon-black-white-outline-clipart-914.jpg"
),
tempfile(fileext = ".jpg")
)We can read the file into R with {magick}.
img <-
list.files(tempdir(), pattern = ".jpg$", full.names = TRUE) |>
magick::image_read()
img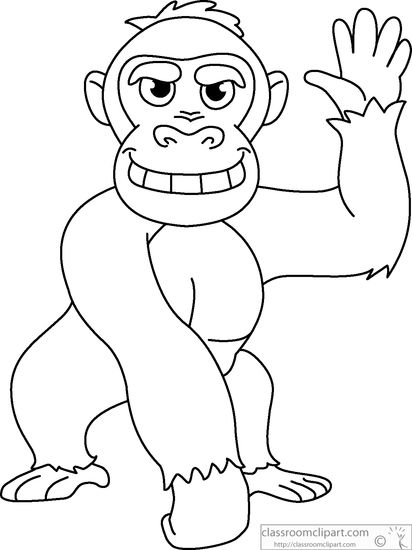
With other {magick} functions we can:
- reduce to two distinct colours only (i.e. for the lines and background), which makes it easier to filter the data later
- convert from an image to point data
go <- img |>
magick::image_quantize(2) |> # colour reduction
magick::image_raster() |> # as x-y data
as.data.frame()
head(go)## x y col
## 1 1 1 #fefefeff
## 2 2 1 #fefefeff
## 3 3 1 #fefefeff
## 4 4 1 #fefefeff
## 5 5 1 #fefefeff
## 6 6 1 #fefefeffAnd to prove we only have two colours (off-white for the background, grey for the lines):
unique(go$col)## [1] "#fefefeff" "#555555ff"Now we can:
- reverse the order of the
yvalues so the gorilla faces the same way as in the paper - filter to retain only the datapoints that represent lines
- rescale the
xandyto create ‘Body Mass Index’ (BMI)1 and ‘steps’ variables
go$y <- rev(go$y)
go <- go[go$col != "#fefefeff", ]
go$bmi <- go$y / max(go $y) * 17 + 15
go$steps <- 15000 - go$x * 15000 / max(go$x)
head(go)## x y col bmi steps
## 174 174 550 #555555ff 32 8665.049
## 175 175 550 #555555ff 32 8628.641
## 176 176 550 #555555ff 32 8592.233
## 196 196 550 #555555ff 32 7864.078
## 198 198 550 #555555ff 32 7791.262
## 199 199 550 #555555ff 32 7754.854You may have noticed that the image has a watermark. We could have removed it earlier with {magick}, but can do it now by filtering out the datapoints in that corner.
go$logo <- ifelse(go$bmi < 16 & go$steps < 5500, TRUE, FALSE)
go <- go[!go$logo, ]This leaves us with 16865 datapoints. We can follow the original study by taking a sample and splitting the results into ‘female’ and ‘male’ groups, weighted so that the female group has higher step counts.
go_smp <- go[sample(nrow(go), 1768), ]
go_smp$rnorm <- rnorm(nrow(go_smp), mean = 0, sd = 10)
go_smp$index <- go_smp$steps * (1 + go_smp$rnorm)
go_smp$group <-
ifelse(go_smp$index < median(go_smp$steps), "F", "M") |>
as.factor()
head(go_smp[, c("bmi", "steps", "group")])## bmi steps group
## 141108 21.42909 7572.81553 F
## 78949 26.09636 5643.20388 F
## 28681 29.86727 5788.83495 M
## 90859 25.20000 7026.69903 F
## 78506 26.12727 6771.84466 M
## 81986 25.88000 72.81553 FNow finally to plot the datasets side-by-side.
par(mfrow = c(1, 2))
with(
go_smp[go_smp$group == "F", ],
plot(
steps, bmi, xlim = c(0, 15000),
pch = 16, cex = 0.5, col = "blue",
xlab = "Steps", ylab = "BMI",
)
)
with(
go_smp[go_smp$group == "M", ],
plot(
steps, bmi, xlim = c(0, 15000),
pch = 16, cex = 0.5, col = "red",
xlab = "Steps", ylab = "BMI"
)
)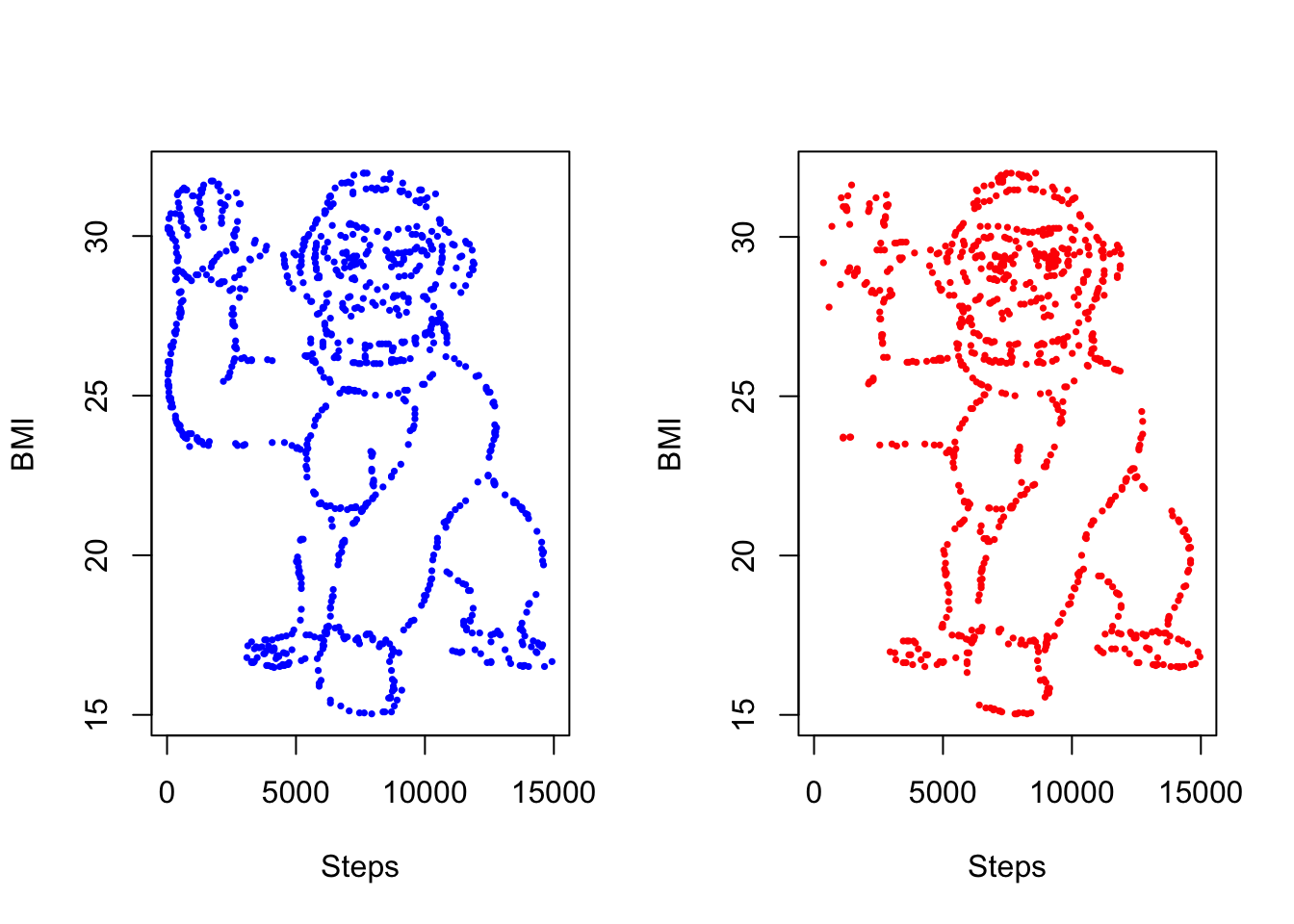
I see them!
This has been a bit overengineered and could be generalised, but it gives a gist of how you might go about converting an image to a dataframe of x and y positions.
At worst, this is a reminder not to trust researchers and to always check for unexpected gorillas.
Session info
## ─ Session info ───────────────────────────────────────────────────────────────
## setting value
## version R version 4.1.1 (2021-08-10)
## os macOS Mojave 10.14.6
## system x86_64, darwin17.0
## ui X11
## language (EN)
## collate en_GB.UTF-8
## ctype en_GB.UTF-8
## tz Europe/London
## date 2021-10-05
##
## ─ Packages ───────────────────────────────────────────────────────────────────
## package * version date lib source
## blogdown 1.5 2021-09-02 [1] CRAN (R 4.1.0)
## bookdown 0.24 2021-09-02 [1] CRAN (R 4.1.0)
## bslib 0.3.0 2021-09-02 [1] CRAN (R 4.1.0)
## cli 3.0.1 2021-07-17 [1] CRAN (R 4.1.0)
## digest 0.6.28 2021-09-23 [1] CRAN (R 4.1.0)
## evaluate 0.14 2019-05-28 [1] CRAN (R 4.1.0)
## fastmap 1.1.0 2021-01-25 [1] CRAN (R 4.1.0)
## highr 0.9 2021-04-16 [1] CRAN (R 4.1.0)
## htmltools 0.5.2 2021-08-25 [1] CRAN (R 4.1.0)
## jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.1.0)
## jsonlite 1.7.2 2020-12-09 [1] CRAN (R 4.1.0)
## knitr 1.34 2021-09-09 [1] CRAN (R 4.1.0)
## magick 2.7.3 2021-08-18 [1] CRAN (R 4.1.0)
## magrittr 2.0.1 2020-11-17 [1] CRAN (R 4.1.0)
## R6 2.5.1 2021-08-19 [1] CRAN (R 4.1.0)
## Rcpp 1.0.7 2021-07-07 [1] CRAN (R 4.1.0)
## rlang 0.4.11 2021-04-30 [1] CRAN (R 4.1.0)
## rmarkdown 2.11 2021-09-14 [1] CRAN (R 4.1.0)
## rstudioapi 0.13 2020-11-12 [1] CRAN (R 4.1.0)
## sass 0.4.0 2021-05-12 [1] CRAN (R 4.1.0)
## sessioninfo 1.1.1 2018-11-05 [1] CRAN (R 4.1.0)
## stringi 1.7.4 2021-08-25 [1] CRAN (R 4.1.0)
## stringr 1.4.0 2019-02-10 [1] CRAN (R 4.1.0)
## withr 2.4.2 2021-04-18 [1] CRAN (R 4.1.0)
## xfun 0.26 2021-09-14 [1] CRAN (R 4.1.0)
## yaml 2.2.1 2020-02-01 [1] CRAN (R 4.1.0)
##
## [1] /Users/matt.dray/Library/R/x86_64/4.1/library
## [2] /Library/Frameworks/R.framework/Versions/4.1/Resources/libraryCheck out a recent episode of the Maintenance Phase podcast (dated 2021-08-03) about the troublesome history and development of BMI as a metric.↩︎